Scripps Research Institute: SARS-CoV-2 and the D614G Mutation
#15,320
All viruses mutate over time, and for the most part, these mutations do little or nothing to affect the function or biological fitness of the virus. It's part of the evolutionary process, and despite Hollywood tropes, mutations are not necessarily ominous or bad.
Single-stranded RNA viruses - like influenza and coronaviruses - are particularly prone to `duplication errors' during replication, and are constantly introducing mutations (see Mechanisms of Viral Mutation).
Most of these mutations end up being of little consequence, and do nothing to affect the transmissibility, replication, host range, or virulence of the virus. Some even prove detrimental, making the virus less `fit' than its predecessors, or attenuating its virulence.
But the reality is, we are in a pandemic because a (likely) bat-borne coronavirus hit the jackpot with the `right' combination of mutations to allow it to jump species, much in the way that a swine-origin H1N1 virus jumped to humans in 2009 to spark the last pandemic.
And as we saw with H1N1pdm, once that virus began to circulate in humans, a small number of worrisome mutations began to appear (see EID Journal: Emergence of D225G Variant A/H1N1, 2013–14 Flu Season, Florida) that affected its virulence.
Luckily, none of the mutations became `fixed', and so they were relatively rare events.
Over the past couple of months we've been watching a curious, and concerning, difference in the spread of COVID-19 in Europe, the United States and South America when compared to China, Vietnam, and other Asian countries.
This has led to speculation that a more transmissible `European' strain had emerged, and was then spread to North and South America.
While many scientists (and public officials) have downplayed the idea, we've seen a number of pre-print studies (see More COVID-19 (SARS-CoV-2) Mutation Reports) that identified a specific mutation (D614G) in the spike protein that appears to enhance its transmissibility.
Among them, this one from the Los Alamos National Laboratory, which describes a new, now dominant `European' strain of COVID-19.
Another study from the University College London, called Emergence of genomic diversity and recurrent mutations in SARS-CoV-2, `. . . . identified close to 200 recurrent genetic mutations in the virus, highlighting how it may be adapting and evolving to its human hosts.'
Yesterday the Scripps Research Institute published a press release on recent findings that seem to support the notion of a `more transmissible' strain of SARS-CoV-2 - originating in Europe - and now also dominant in North and South America.
Their study - still under peer review - can be viewed at:
While this study adds further weight to the idea of a `more transmissible strain' SARS-CoV-2, more work will be needed to confirm the theory. The press release follows:
The mutation had the effect of markedly increasing the number of functional spikes on the viral surface, she adds. Those spikes are what allow the virus to bind to and infect cells.
“The number—or density—of functional spikes on the virus is 4 or 5 times greater due to this mutation,” Choe says.
More flexible spikes allow newly made viral particles to navigate the journey from producer cell to target cell fully intact, with less tendency to fall apart prematurely, he explains.
“Our data are very clear, the virus becomes much more stable with the mutation,” Choe says.
There has been much debate about why COVID-19 outbreaks in Italy and New York have so quickly overwhelmed health systems, while early outbreaks in places like San Francisco and Washington state proved more readily managed, at least initially. Was it something about those communities and their response, or had the virus somehow changed?
All viruses acquire minute genetic changes as they reproduce and spread. Those changes rarely impact fitness or ability to compete. The SARS-CoV-2 variant that circulated in the earliest regional outbreaks lacked the D614G mutation now dominating in much of the world.
But was that because of the so-called “founder effect,” seen when a small number of variants fan out into a wide population, by chance? Choe and Farzan believe their biochemical experiments settle the question.
Choe and Farzan’s paper is titled “The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity.” Now undergoing peer review, it is being posted prior to publication to the pre-print site bioRxiv, and released early, amid news reports of its findings.
Choe and Farzan note that their research was performed using harmless viruses engineered to produce key coronavirus proteins. Whether the changes they observed also translate to increased transmissibility in the real world requires additional epidemiological studies, they note.
Encouragingly, the duo found that immune factors from the serum of infected people work equally well against engineered viruses both with and without the D614G mutation. That’s a hopeful sign that vaccine candidates in development will work against variants with or without that mutation, Choe says.
Choe and Farzan have studied coronaviruses for nearly 20 years, since the first outbreak of SARS, a similar virus. They were the first to discover in 2003 that SARS bound to the ACE2 receptor on cells. Others’ experiments have shown the SARS-CoV-2 virus binds the same ACE2 receptor.
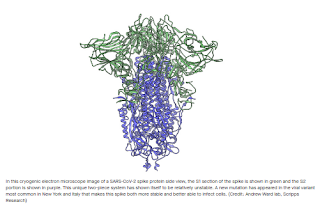
But Farzan and Choe note a key structural difference between spike proteins on the first SARS virus and this new pandemic strain. With both, under an electron microscope, the spike has tripod shape, with its three segments bound together at a backbone-like scaffold. But SARS-CoV-2 is different. Its tripod is divided in two discreet segments, S1 and S2.
Initially, this unusual feature produced unstable spikes, Farzan says. Only about a quarter of the hundreds of spikes on each SARS-CoV-2 virus maintain the structure they need to successfully infect a target cell. With the mutation, the tripod breaks much less frequently, meaning more of its spikes are fully functional, he says.
The addition of the D614G mutation means that the amino acid at that location is switched from aspartic acid to glycine. That renders it more bendable, Farzan says.
Evidence of its success can be seen in the sequenced strains that scientists globally are contributing to databases including GenBank, the duo reports. In February, no sequences deposited to the GenBank database showed the D614G mutation. But by March, it appeared in 1 out of 4 samples. In May, it appeared in 70 percent of samples, Farzan says.
Over time, it has figured out how to hold on better and not fall apart until it needs to,” Farzan says. “The virus has, under selection pressure, made itself more stable.”
It is still unknown whether this small mutation affects the severity of symptoms of infected people, or increases mortality, the scientists say. While ICU data from New York and elsewhere reports a preponderance of the new D614G variant, much more data, ideally under controlled studies, are needed, Choe says.
In addition to senior authors Choe and Farzan, the authors include first authors Lizhou Zhang, Cody Jackson and Huihui Mou, plus co-authors Amrita Ojha, Erumbi Rangarajan and Tina Izard, all of Scripps Research.
The work was supported by the National Institutes of Health through an administrative supplement to RO1 AI129868 for coronavirus research.
https://afludiary.blogspot.com/2020/...ars-cov-2.html

#15,320
All viruses mutate over time, and for the most part, these mutations do little or nothing to affect the function or biological fitness of the virus. It's part of the evolutionary process, and despite Hollywood tropes, mutations are not necessarily ominous or bad.
Single-stranded RNA viruses - like influenza and coronaviruses - are particularly prone to `duplication errors' during replication, and are constantly introducing mutations (see Mechanisms of Viral Mutation).
Most of these mutations end up being of little consequence, and do nothing to affect the transmissibility, replication, host range, or virulence of the virus. Some even prove detrimental, making the virus less `fit' than its predecessors, or attenuating its virulence.
But the reality is, we are in a pandemic because a (likely) bat-borne coronavirus hit the jackpot with the `right' combination of mutations to allow it to jump species, much in the way that a swine-origin H1N1 virus jumped to humans in 2009 to spark the last pandemic.
And as we saw with H1N1pdm, once that virus began to circulate in humans, a small number of worrisome mutations began to appear (see EID Journal: Emergence of D225G Variant A/H1N1, 2013–14 Flu Season, Florida) that affected its virulence.
Luckily, none of the mutations became `fixed', and so they were relatively rare events.
Over the past couple of months we've been watching a curious, and concerning, difference in the spread of COVID-19 in Europe, the United States and South America when compared to China, Vietnam, and other Asian countries.
This has led to speculation that a more transmissible `European' strain had emerged, and was then spread to North and South America.
While many scientists (and public officials) have downplayed the idea, we've seen a number of pre-print studies (see More COVID-19 (SARS-CoV-2) Mutation Reports) that identified a specific mutation (D614G) in the spike protein that appears to enhance its transmissibility.
Among them, this one from the Los Alamos National Laboratory, which describes a new, now dominant `European' strain of COVID-19.
B Korber, WM Fischer, S Gnanakaran, H Yoon, J Theiler, W Abfalterer, B Foley, EE Giorgi, T Bhattacharya, MD Parker, DG Partridge, CM Evans, TI de Silva, on behalf of the Sheffield COVID-19 Genomics Group, CC LaBranche, DC Montefiori
Another study from the University College London, called Emergence of genomic diversity and recurrent mutations in SARS-CoV-2, `. . . . identified close to 200 recurrent genetic mutations in the virus, highlighting how it may be adapting and evolving to its human hosts.'
Yesterday the Scripps Research Institute published a press release on recent findings that seem to support the notion of a `more transmissible' strain of SARS-CoV-2 - originating in Europe - and now also dominant in North and South America.
Their study - still under peer review - can be viewed at:
Lizhou Zhang1#, Cody B Jackson1#, Huihui Mou1#, Amrita Ojha1, Erumbi S Rangarajan2, Tina Izard2, Michael Farzan1*, Hyeryun Choe1*
While this study adds further weight to the idea of a `more transmissible strain' SARS-CoV-2, more work will be needed to confirm the theory. The press release follows:
COVID-19-causing viral variant taking over in the United States and Europe now carries more functional, cell-binding spikes.
June 12, 2020
Additional Resources
Additional Resources
JUPITER, FL — A tiny genetic mutation in the SARS coronavirus 2 variant circulating throughout Europe and the United States significantly increases the virus’ ability to infect cells, lab experiments performed at Scripps Research show.
“Viruses with this mutation were much more infectious than those without the mutation in the cell culture system we used,” says Scripps
Research virologist Hyeryun Choe, PhD, senior author of the study.
Research virologist Hyeryun Choe, PhD, senior author of the study.
The mutation had the effect of markedly increasing the number of functional spikes on the viral surface, she adds. Those spikes are what allow the virus to bind to and infect cells.
“The number—or density—of functional spikes on the virus is 4 or 5 times greater due to this mutation,” Choe says.
The spikes give the coronavirus its crown-like appearance and enable it to latch onto target cell receptors called ACE2. The mutation, called D614G, provides greater flexibility to the spike’s “backbone,” explains co-author Michael Farzan, PhD, co-chairman of the Scripps Research Department of Immunology and Microbiology.
More flexible spikes allow newly made viral particles to navigate the journey from producer cell to target cell fully intact, with less tendency to fall apart prematurely, he explains.
“Our data are very clear, the virus becomes much more stable with the mutation,” Choe says.
There has been much debate about why COVID-19 outbreaks in Italy and New York have so quickly overwhelmed health systems, while early outbreaks in places like San Francisco and Washington state proved more readily managed, at least initially. Was it something about those communities and their response, or had the virus somehow changed?
All viruses acquire minute genetic changes as they reproduce and spread. Those changes rarely impact fitness or ability to compete. The SARS-CoV-2 variant that circulated in the earliest regional outbreaks lacked the D614G mutation now dominating in much of the world.
But was that because of the so-called “founder effect,” seen when a small number of variants fan out into a wide population, by chance? Choe and Farzan believe their biochemical experiments settle the question.
“
There have been at least a dozen scientific papers talking about the predominance of this mutation,” Farzan says. “Are we just seeing a ‘founder effect?’ Our data nails it. It is not the founder effect.”
There have been at least a dozen scientific papers talking about the predominance of this mutation,” Farzan says. “Are we just seeing a ‘founder effect?’ Our data nails it. It is not the founder effect.”
Choe and Farzan’s paper is titled “The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity.” Now undergoing peer review, it is being posted prior to publication to the pre-print site bioRxiv, and released early, amid news reports of its findings.
Choe and Farzan note that their research was performed using harmless viruses engineered to produce key coronavirus proteins. Whether the changes they observed also translate to increased transmissibility in the real world requires additional epidemiological studies, they note.
Encouragingly, the duo found that immune factors from the serum of infected people work equally well against engineered viruses both with and without the D614G mutation. That’s a hopeful sign that vaccine candidates in development will work against variants with or without that mutation, Choe says.
Choe and Farzan have studied coronaviruses for nearly 20 years, since the first outbreak of SARS, a similar virus. They were the first to discover in 2003 that SARS bound to the ACE2 receptor on cells. Others’ experiments have shown the SARS-CoV-2 virus binds the same ACE2 receptor.
But Farzan and Choe note a key structural difference between spike proteins on the first SARS virus and this new pandemic strain. With both, under an electron microscope, the spike has tripod shape, with its three segments bound together at a backbone-like scaffold. But SARS-CoV-2 is different. Its tripod is divided in two discreet segments, S1 and S2.
Initially, this unusual feature produced unstable spikes, Farzan says. Only about a quarter of the hundreds of spikes on each SARS-CoV-2 virus maintain the structure they need to successfully infect a target cell. With the mutation, the tripod breaks much less frequently, meaning more of its spikes are fully functional, he says.
The addition of the D614G mutation means that the amino acid at that location is switched from aspartic acid to glycine. That renders it more bendable, Farzan says.
Evidence of its success can be seen in the sequenced strains that scientists globally are contributing to databases including GenBank, the duo reports. In February, no sequences deposited to the GenBank database showed the D614G mutation. But by March, it appeared in 1 out of 4 samples. In May, it appeared in 70 percent of samples, Farzan says.
Over time, it has figured out how to hold on better and not fall apart until it needs to,” Farzan says. “The virus has, under selection pressure, made itself more stable.”
It is still unknown whether this small mutation affects the severity of symptoms of infected people, or increases mortality, the scientists say. While ICU data from New York and elsewhere reports a preponderance of the new D614G variant, much more data, ideally under controlled studies, are needed, Choe says.
In addition to senior authors Choe and Farzan, the authors include first authors Lizhou Zhang, Cody Jackson and Huihui Mou, plus co-authors Amrita Ojha, Erumbi Rangarajan and Tina Izard, all of Scripps Research.
The work was supported by the National Institutes of Health through an administrative supplement to RO1 AI129868 for coronavirus research.
https://afludiary.blogspot.com/2020/...ars-cov-2.html
Comment