Re: 225G Preliminary Worldwide Tracking & Evaluation
hat tip FT Resident SusanC -
<table width="100%"><tbody><tr><td> Receptor Binding and Evolutionary Trends of H1N1 HAs
by: SusanC
Sat Nov 21, 2009 at 23:28:40 PM EST
</td> </tr> <tr> <td>
</td> </tr> <tr> <td class="theFlip"> SusanC :: Receptor Binding and Evolutionary Trends of H1N1 HAs</td> </tr> <tr> <td>There's been much interest recently in the revelation of 'mutations' in the HA of the 2009 H1N1 virus, specifically, a 'change' at position 225 from D to G (see codon table for coding). I put the words mutation and change in quotes for reasons that will become obvious as you read further. I thought I might as well take this opportunity to review the issue of mutations that favor different patterns of receptor binding, from 1918 to now. The bulk of the following discussion is based on 2 papers:
Here's a quote from Stevens et al that summarizes, in a much simplified form, the issue of receptor binding:
Second, from Shen et al, just published:
<center> </center>
</center>
COMMENTS, on position 225:
Receptor binding is by no means the only or even the most important driver of virulence. In the 2003-04 flu season, a new subclade of H3N2 emerged which was very virulent and caused much higher fatality (as discussed here). This virus had exactly the same HA as one that had been circulating before as a minor (ie non-dominant) strain, the only differences are in the polymerase genes. Yet, whereas the previous version caused mild disease and was not able to wipe out other strains, the new one (with exactly the same HA) both became dominant and was much more virulent.
See Memoli 2009 Recent human influenza A/H3N2 virus evolution driven by novel selection factors in addition to antigenic drift. and my comments here and here
</td></tr></tbody></table>
hat tip FT Resident SusanC -
<table width="100%"><tbody><tr><td> Receptor Binding and Evolutionary Trends of H1N1 HAs
by: SusanC
Sat Nov 21, 2009 at 23:28:40 PM EST
</td> </tr> <tr> <td>
</td> </tr> <tr> <td class="theFlip"> SusanC :: Receptor Binding and Evolutionary Trends of H1N1 HAs</td> </tr> <tr> <td>There's been much interest recently in the revelation of 'mutations' in the HA of the 2009 H1N1 virus, specifically, a 'change' at position 225 from D to G (see codon table for coding). I put the words mutation and change in quotes for reasons that will become obvious as you read further. I thought I might as well take this opportunity to review the issue of mutations that favor different patterns of receptor binding, from 1918 to now. The bulk of the following discussion is based on 2 papers:
- Stevens 2006 Glycan Microarray Analysis of the Hemagglutinins from Modern and Pandemic Influenza Viruses Reveals Different Receptor Specificities
- Shen 2009 Evolutionary Trends of A(H1N1) Influenza Virus Hemagglutinin Since 1918.
Here's a quote from Stevens et al that summarizes, in a much simplified form, the issue of receptor binding:
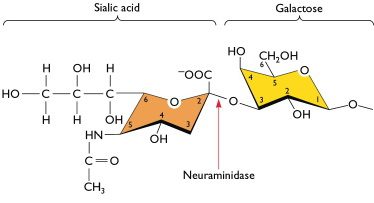
Receptor binding specificity is primarily distinguished by recognition of the terminal sialic acid and its linkage to the vicinal galactose of carbohydrates in these tissues. Avian viruses preferentially bind to receptors with an a2-3 linkage, whereas human adapted viruses are specific for the a2-6 linkage.3,4 The subtle switch from a2-3 to a2-6 receptor specificity is thought to be a critical step in the adaptation of avian viruses to a human host.
First, findings from Stevens et al: - Using glycan microarray (ie binding to sugars on slides, basically), they found 2 positions where changes resulted in shifting of receptor binding preference, 190 and 225
- In position 190, avian viruses had E, and viruses from humans (including 1918) had a D. The former binds to a2,3 sites, the latter binds to a2,6 sites. Very straight-forward. One single change completely changes the receptor binding from avian to human.
- For position 225, in 1918, there were 2 variants, D and G. These 2 existed both early and late in the pandemic. The 2 sequences tested in this paper were from 2 soldiers who died on the same day, one in NY, one in South Carolina. The SC one had HAs with a D in position 225, the NY one had a G.
- In the glycan microarray test, the SC HA (with D at 190 and D at 225), bound exclusively to a2,6 (ie preferential for human), whereas the NY HA (with D at 190 and G at 225) had mixed specificity, for both a2,3 and a2,6 (both avian and human)
- However, the authors noted that despite these differences in receptor binding preference in the lab, these 2 variants showed no difference in virulence or transmissibility
Second, from Shen et al, just published:
<center>
 </center>
</center> - During a pandemic, in 1918 and in 2009, position 190 is strictly conserved, ie 100% D.
- In interpandemic years, there is some selection pressure on position 190, away from D (to what, they didn't say. Or at least I didn't find it).
- For position 225, during a pandemic, in both 1918 and 2009, both variants are present.
- In interpandemic years, in samples that have been passaged through eggs (ie egg-adapted) there is selection pressure from D to G.
- For interpandemic samples that are not egg-adapted (from 1979-2008), 98.2% of samples had D, but changes in this position did not fulfill their criteria for having been under selection pressure. Nevertheless, D at 225 is highly conserved, in interpandemic years.
COMMENTS, on position 225:
- The 2 variants in position 225, co-existed in 1918. We are now finding the same variants in 2009.
- In between pandemics, ie after the virus has jumped to humans, position 225 tends to become highly conserved, as D.
- There is no evidence of evolutionary pressure (ie 'change') from D to G, either in 1918, in 2009, or in all the years in between. If one were to infer any selection pressure, it would be from G to D, since in samples that are not egg-adapted, D was almost exclusively found, eg 1979-2008.
- In any case, a D225G change in 1918 resulted in lower preference to human receptors. (see Stevens et al, above)
- And, in 1918, these 2 variants were not associated with differences in virulence or transmissibility. As far as we know, that appears to be the case as well, in 2009, since both variants have been found from the earliest days of this pandemic, since April, and in many countries. (more here)
Receptor binding is by no means the only or even the most important driver of virulence. In the 2003-04 flu season, a new subclade of H3N2 emerged which was very virulent and caused much higher fatality (as discussed here). This virus had exactly the same HA as one that had been circulating before as a minor (ie non-dominant) strain, the only differences are in the polymerase genes. Yet, whereas the previous version caused mild disease and was not able to wipe out other strains, the new one (with exactly the same HA) both became dominant and was much more virulent.
See Memoli 2009 Recent human influenza A/H3N2 virus evolution driven by novel selection factors in addition to antigenic drift. and my comments here and here
</td></tr></tbody></table>

Comment