Preprint: The Continuous Evolution of SARS-CoV-2 in South Africa (Lineage C.1.2)
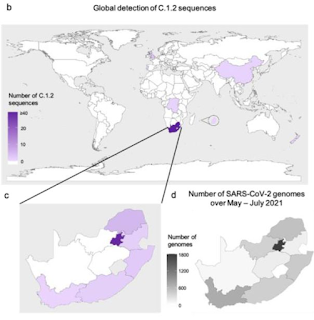
Global Detection of C.1.2 Sequences
#16,153
Although thousands of COVID variants have been detected around the world, only a dozen or so have been classified as VOIs (Variants of Interest), and fewer still are considered VOCs (Variants of Concern).
Some VOIs, after initially raising concerns - have already gone extinct - unable to successfully compete against more aggressive variants like Delta. What may look worrisome today, could become a brief footnote in the COVID pandemic's history a few months from now.
The COVID viral landscape is highly dynamic, continually changing, and its future is very, very hard to predict. Last April when Alpha was on the ascendent world-wide, it was very hard to see it being supplanted within months by an upstart Delta variant coming out of India.
But that's exactly what happened.
With 99% of the COVID cases in the United States estimated to be from Delta, it is difficult to envision that variant being displaced anytime soon by something else, yet that possibility exists. So researcher are on the lookout for potential contenders; variants that may have the ability to compete head-to-head with Delta.
Last month's rising star was thought to be Lambda (see PrePrint: Infectivity and Immune Escape of the New SARS-CoV-2 Variant of Interest Lambda), with pockets of Beta, Gamma, and Epsilon all still in the running.
Unless and until Delta relinquishes its stranglehold, however, identifying a likely successor is nearly impossible.
There are too many variables and interactions we don't fully understand, and always the possibility that a `new' and more `biologically fit' variant will emerge and join the fray.
To that end we have a preprint which identifies a `highly mutated' COVID variant in South Africa (C.1.2) , that carries a large number of `red flag' mutations which have been shown to increase biological fitness in VOCs and VOIs.
This preprint has gotten a lot of notice over the weekend, both on social media and in news media reports, including:
New variant, C.1.2, may be more infectious, evade vaccine protection: Study
New ‘most mutated COVID-19 variant’ found in South Africa
New COVID variant detected in South Africa, most mutated variant so far
Alarming headlines aside, the actual number of C.1.2 variants discovered to date is small (n < 100), and its geographic spread is limited. That said, C.1.2 is most closely related to the Lambda variant - which has gained its own notoriety - and it seems to be evolving at a much faster clip than other variants.
Both of which make this a variant worth exploring. But whether that means C.1.2 will become the next `big challenge' in COVID, is far from clear.
C.1.2 lineage - which is a highly mutated form of C.1 first reported last January - was first detected in the Mpumalanga and Gauteng provinces of South Africa in May. It has since been detected across much of South Africa, as well as sporadic cases in England, China, and parts of Europe and Oceania.

https://cov-lineages.org/lineage_list.html
Whether or not C.1.2 is the heir to Delta's throne, or another flash in the pan, is unknown. But it does serve as warning that not all variants evolve at the same rate, and that there are potential `wildcards' in play in this pandemic.
While our immediate problem is dealing with Delta, it doesn't hurt to look ahead, as long as we don't focus too heavily on any one particular outcome.
A link, and the abstract, from the preprint follow. Those who don't mind slugging through some fairly technical text will want to read the preprint in its entirety. I'll have a brief postscript when you return.
The continuous evolution of SARS-CoV-2 in South Africa: a new lineage with rapid accumulation of mutations of concern and global detection
Cathrine Scheepers, Josie Everatt, Daniel G. Amoako, Anele Mnguni, Arshad Ismail, Boitshoko Mahlangu, Constantinos Kurt Wibmer, Eduan Wilkinson, Houriiyah Tegally, James Emmanuel San, Jennifer Giandhari, Noxolo Ntuli, Sureshnee Pillay, Thabo Mohale, Yeshnee Naidoo, Zamantungwa T. Khumalo, Zinhle Makatini, NGS-SA, Alex Sigal, Carolyn Williamson, Florette Treurnicht, Koleka Mlisana, Marietjie Venter, Nei-yuan Hsiao, Nicole Wolter, Nokukhanya Msomi, Richard Lessells, Tongai Maponga, Wolfgang Preiser, Penny L. Moore, Anne von Gottberg, Tulio de Oliveira, Jinal N. Bhiman
(Continue . . . )
The authors suggest hat the rapid accumulation of mutations in C.1.2 may have occurred in a single, immunocompromised, individual who could have carried the virus for a prolonged period of time. This is not the first time we've seen this notion.
Last December, the ECDC Threat Assessment Brief On UK SARS-CoV-2 Variant speculated that the large number of concurrent mutations in the B.1.1.7 variant may have arised from a single chronically-infected, likely immunocompromised, individual.
About the same time, COG-UK released a report on multiple escape mutants generated in a chronically ill, immunocompromised patient, after receiving convalescent plasma therapy. Three weeks ago the NEJM carried an article warning of the potential risk of new variants emerging from long-term infections among immunocompromised individuals (see SARS-CoV-2 Variants in Patients with Immunosuppression), a concern we've seen raised previously.
Below you'll find a link to the press release from the Fred Hutchinson Cancer Research Center on their NEJM article mentioned above.
Earlier this month, in COCA Call Tomorrow (Aug 12th): Therapeutic Options to Prevent Severe COVID-19 in Immunocompromised People, we looked at the importance preventing infection in immunocompromised individuals whenever possible.
Regardless of how C.1.2 jumped ahead in the mutation race, this is a reminder that COVID is perfectly capable of surprising us, and that this pandemic may still have some unexpected twists and turns ahead.

Global Detection of C.1.2 Sequences
#16,153
Although thousands of COVID variants have been detected around the world, only a dozen or so have been classified as VOIs (Variants of Interest), and fewer still are considered VOCs (Variants of Concern).
Some VOIs, after initially raising concerns - have already gone extinct - unable to successfully compete against more aggressive variants like Delta. What may look worrisome today, could become a brief footnote in the COVID pandemic's history a few months from now.
The COVID viral landscape is highly dynamic, continually changing, and its future is very, very hard to predict. Last April when Alpha was on the ascendent world-wide, it was very hard to see it being supplanted within months by an upstart Delta variant coming out of India.
But that's exactly what happened.
With 99% of the COVID cases in the United States estimated to be from Delta, it is difficult to envision that variant being displaced anytime soon by something else, yet that possibility exists. So researcher are on the lookout for potential contenders; variants that may have the ability to compete head-to-head with Delta.
Last month's rising star was thought to be Lambda (see PrePrint: Infectivity and Immune Escape of the New SARS-CoV-2 Variant of Interest Lambda), with pockets of Beta, Gamma, and Epsilon all still in the running.
Unless and until Delta relinquishes its stranglehold, however, identifying a likely successor is nearly impossible.
There are too many variables and interactions we don't fully understand, and always the possibility that a `new' and more `biologically fit' variant will emerge and join the fray.
To that end we have a preprint which identifies a `highly mutated' COVID variant in South Africa (C.1.2) , that carries a large number of `red flag' mutations which have been shown to increase biological fitness in VOCs and VOIs.
This preprint has gotten a lot of notice over the weekend, both on social media and in news media reports, including:
New variant, C.1.2, may be more infectious, evade vaccine protection: Study
New ‘most mutated COVID-19 variant’ found in South Africa
New COVID variant detected in South Africa, most mutated variant so far
Alarming headlines aside, the actual number of C.1.2 variants discovered to date is small (n < 100), and its geographic spread is limited. That said, C.1.2 is most closely related to the Lambda variant - which has gained its own notoriety - and it seems to be evolving at a much faster clip than other variants.
Both of which make this a variant worth exploring. But whether that means C.1.2 will become the next `big challenge' in COVID, is far from clear.
C.1.2 lineage - which is a highly mutated form of C.1 first reported last January - was first detected in the Mpumalanga and Gauteng provinces of South Africa in May. It has since been detected across much of South Africa, as well as sporadic cases in England, China, and parts of Europe and Oceania.

https://cov-lineages.org/lineage_list.html
Whether or not C.1.2 is the heir to Delta's throne, or another flash in the pan, is unknown. But it does serve as warning that not all variants evolve at the same rate, and that there are potential `wildcards' in play in this pandemic.
While our immediate problem is dealing with Delta, it doesn't hurt to look ahead, as long as we don't focus too heavily on any one particular outcome.
A link, and the abstract, from the preprint follow. Those who don't mind slugging through some fairly technical text will want to read the preprint in its entirety. I'll have a brief postscript when you return.
The continuous evolution of SARS-CoV-2 in South Africa: a new lineage with rapid accumulation of mutations of concern and global detection
Cathrine Scheepers, Josie Everatt, Daniel G. Amoako, Anele Mnguni, Arshad Ismail, Boitshoko Mahlangu, Constantinos Kurt Wibmer, Eduan Wilkinson, Houriiyah Tegally, James Emmanuel San, Jennifer Giandhari, Noxolo Ntuli, Sureshnee Pillay, Thabo Mohale, Yeshnee Naidoo, Zamantungwa T. Khumalo, Zinhle Makatini, NGS-SA, Alex Sigal, Carolyn Williamson, Florette Treurnicht, Koleka Mlisana, Marietjie Venter, Nei-yuan Hsiao, Nicole Wolter, Nokukhanya Msomi, Richard Lessells, Tongai Maponga, Wolfgang Preiser, Penny L. Moore, Anne von Gottberg, Tulio de Oliveira, Jinal N. Bhiman
Abstract
SARS-CoV-2 variants of interest have been associated with increased transmissibility, neutralization resistance and disease severity. Ongoing SARS-CoV-2 genomic surveillance world-wide has improved our ability to rapidly identify such variants. Here we report the identification of a potential variant of interest assigned to the PANGO lineage C.1.2. This lineage was first identified in May 2021 and evolved from C.1, one of the lineages that dominated the first wave of SARS-CoV-2 infections in South Africa and was last detected in January 2021. C.1.2 has since been detected across the majority of the provinces in South Africa and in seven other countries spanning Africa, Europe, Asia and Oceania.
The emergence of C.1.2 was associated with an increased substitution rate, as was previously observed with the emergence of the Alpha, Beta and Gamma variants of concern (VOCs). C.1.2 contains multiple substitutions (R190S, D215G, E484K, N501Y, H655Y and T859N) and deletions (Y144del, L242-A243del) within the spike protein, which have been observed in other VOCs and are associated with increased transmissibility and reduced neutralization sensitivity. Of greater concern is the accumulation of additional mutations (C136F, Y449H and N679K) which are also likely to impact neutralization sensitivity or furin cleavage and therefore replicative fitness. While the phenotypic characteristics and epidemiology of C.1.2 are being defined, it is important to highlight this lineage given its concerning constellations of mutations.
(SNIP)
Discussion/Conclusion
We have identified a new SARS-CoV-2 variant assigned to the PANGO lineage C.1.2. This variant has been detected throughout the third wave of infections in South Africa from May 2021 onwards and has been detected in seven other countries within Europe, Asia, Africa and Oceania. The identification of novel SARS-CoV-2 variants is commonly associated with new waves of infection. Like several other VOCs, C.1.2 has accumulated a number of substitutions beyond what would be expected from the background SARS-CoV-2 evolutionary rate. This suggests the likelihood that these mutations arose during a period of accelerated evolution in a single individual with prolonged viral infection through virus-host co-evolution19–21. Deletions within the NTD (like Y144del, seen in C.1.2 and other VOCs) have been evident in cases of prolonged infection, further supporting this hypothesis22–24.
C.1.2 contains many mutations that have been identified in all four VOCs (Alpha, Beta, Delta and Gamma) and three VOIs (Kappa, Eta and Lambda) as well as additional mutations within the NTD (C136F), RBD (Y449H), and adjacent to the furin cleavage site (N679K). Many of the shared mutations have been associated with improved ACE2 binding (N501Y)25–29 or furin cleavage (H655Y and P681H/R)30–32, and reduced neutralization activity (particularly Y144del, 242-244del, and E484K)17,33–39, providing sufficient cause for concern of continued transmission of this variant. Future work aims to determine the functional impact of these mutations, which likely include neutralizing antibody escape, and to investigate whether their combination confers a replicative fitness advantage over the Delta variant.
The C.1.2 lineage is continuing to grow. At the time of submission (20 August 2021) there were 80 C.1.2 sequences in GISAID with it now having been detected in Botswana and in the Northern Cape of South Africa.
Discussion/Conclusion
We have identified a new SARS-CoV-2 variant assigned to the PANGO lineage C.1.2. This variant has been detected throughout the third wave of infections in South Africa from May 2021 onwards and has been detected in seven other countries within Europe, Asia, Africa and Oceania. The identification of novel SARS-CoV-2 variants is commonly associated with new waves of infection. Like several other VOCs, C.1.2 has accumulated a number of substitutions beyond what would be expected from the background SARS-CoV-2 evolutionary rate. This suggests the likelihood that these mutations arose during a period of accelerated evolution in a single individual with prolonged viral infection through virus-host co-evolution19–21. Deletions within the NTD (like Y144del, seen in C.1.2 and other VOCs) have been evident in cases of prolonged infection, further supporting this hypothesis22–24.
C.1.2 contains many mutations that have been identified in all four VOCs (Alpha, Beta, Delta and Gamma) and three VOIs (Kappa, Eta and Lambda) as well as additional mutations within the NTD (C136F), RBD (Y449H), and adjacent to the furin cleavage site (N679K). Many of the shared mutations have been associated with improved ACE2 binding (N501Y)25–29 or furin cleavage (H655Y and P681H/R)30–32, and reduced neutralization activity (particularly Y144del, 242-244del, and E484K)17,33–39, providing sufficient cause for concern of continued transmission of this variant. Future work aims to determine the functional impact of these mutations, which likely include neutralizing antibody escape, and to investigate whether their combination confers a replicative fitness advantage over the Delta variant.
The C.1.2 lineage is continuing to grow. At the time of submission (20 August 2021) there were 80 C.1.2 sequences in GISAID with it now having been detected in Botswana and in the Northern Cape of South Africa.
(Continue . . . )
The authors suggest hat the rapid accumulation of mutations in C.1.2 may have occurred in a single, immunocompromised, individual who could have carried the virus for a prolonged period of time. This is not the first time we've seen this notion.
Last December, the ECDC Threat Assessment Brief On UK SARS-CoV-2 Variant speculated that the large number of concurrent mutations in the B.1.1.7 variant may have arised from a single chronically-infected, likely immunocompromised, individual.
About the same time, COG-UK released a report on multiple escape mutants generated in a chronically ill, immunocompromised patient, after receiving convalescent plasma therapy. Three weeks ago the NEJM carried an article warning of the potential risk of new variants emerging from long-term infections among immunocompromised individuals (see SARS-CoV-2 Variants in Patients with Immunosuppression), a concern we've seen raised previously.
Below you'll find a link to the press release from the Fred Hutchinson Cancer Research Center on their NEJM article mentioned above.
Earlier this month, in COCA Call Tomorrow (Aug 12th): Therapeutic Options to Prevent Severe COVID-19 in Immunocompromised People, we looked at the importance preventing infection in immunocompromised individuals whenever possible.
You can find more information on the use of monoclonal antibodies in the NIH's August 4th update of their Anti-SARS-CoV-2 Monoclonal Antibodies page.
Regardless of how C.1.2 jumped ahead in the mutation race, this is a reminder that COVID is perfectly capable of surprising us, and that this pandemic may still have some unexpected twists and turns ahead.
