CDC: New Respiratory Bug Has Killed 10
ATLANTA (AP) - A mutated version of a common cold virus has caused 10 deaths in the last 18 months, U.S. health officials said Thursday.
Adenoviruses usually cause respiratory infections that aren't considered lethal. But a new variant has caused at least 140 illnesses in New York, Oregon, Washington and Texas, according to a report issued Thursday by the U.S. Centers for Disease Control and Prevention.
The illness made headlines in Texas earlier this year, when a so- called boot camp flu sickened hundreds at Lackland Air Force Base in San Antonio. The most serious cases were blamed on the emerging virus and one 19-year-old trainee died.
"What really got people's attention is these are healthy young adults landing in the hospital and, in some cases, the ICU," said Dr. John Su, an infectious diseases investigator with the CDC.
There are more than 50 distinct types of adenoviruses tied to human illnesses. They are one cause of the common cold, and also trigger pneumonia and bronchitis. Severe illnesses are more likely in people with weaker immune systems.
Some adenoviruses have also been blamed for gastroenteritis, conjunctivitis and cystitis.
There are no good antiviral medications for adenoviruses. Patients usually are treated with aspirin, liquids and bed rest.
In the CDC report, the earliest case of the mutated virus was found in an infant girl in New York City, who died last year. The child seemed healthy right after birth, but then became dehydrated and lost appetite. She died 12 days after she was born.
Tests found that she been infected with a form of adenovirus, called Ad14, but with some little differences, Su said.
It's not clear how the changes made it more lethal, said Linda Gooding, an Emory University researcher who specializes in adenoviruses.
Earlier this year, hundreds of trainees at Lackland became ill with respiratory infections. Tests showed a variety of adenoviruses in the trainees, but at least 106?and probably more?had the mutated form of Ad14, including five who ended up in an intensive care unit
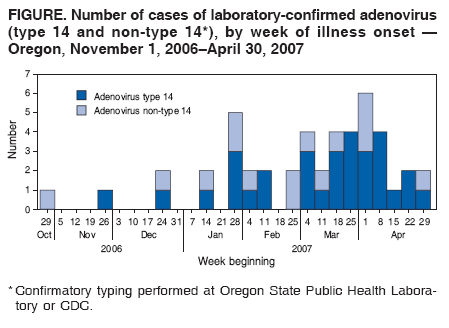
In April, Oregon health officials learned of a cluster of cases at a Portland-area hospital. They ultimately counted 31 cases, including seven who died with severe pneumonia. The next month, Washington state officials reported four hospitalized patients had the same mutated virus. One, who also had AIDS, died.
The Ad14 form of adenovirus was first identified in 1955. In 1969, it was blamed for a rash of illnesses in military recruits stationed in Europe, but it's been detected rarely since then. But it seems to growing more common. The strain accounted for 6 percent of adenovirus samples collected in 22 medical facilities in 2006, while none was seen the previous two years, according to a study published this month in the medical journal Clinical Infectious Diseases.
ATLANTA (AP) - A mutated version of a common cold virus has caused 10 deaths in the last 18 months, U.S. health officials said Thursday.
Adenoviruses usually cause respiratory infections that aren't considered lethal. But a new variant has caused at least 140 illnesses in New York, Oregon, Washington and Texas, according to a report issued Thursday by the U.S. Centers for Disease Control and Prevention.
The illness made headlines in Texas earlier this year, when a so- called boot camp flu sickened hundreds at Lackland Air Force Base in San Antonio. The most serious cases were blamed on the emerging virus and one 19-year-old trainee died.
"What really got people's attention is these are healthy young adults landing in the hospital and, in some cases, the ICU," said Dr. John Su, an infectious diseases investigator with the CDC.
There are more than 50 distinct types of adenoviruses tied to human illnesses. They are one cause of the common cold, and also trigger pneumonia and bronchitis. Severe illnesses are more likely in people with weaker immune systems.
Some adenoviruses have also been blamed for gastroenteritis, conjunctivitis and cystitis.
There are no good antiviral medications for adenoviruses. Patients usually are treated with aspirin, liquids and bed rest.
In the CDC report, the earliest case of the mutated virus was found in an infant girl in New York City, who died last year. The child seemed healthy right after birth, but then became dehydrated and lost appetite. She died 12 days after she was born.
Tests found that she been infected with a form of adenovirus, called Ad14, but with some little differences, Su said.
It's not clear how the changes made it more lethal, said Linda Gooding, an Emory University researcher who specializes in adenoviruses.
Earlier this year, hundreds of trainees at Lackland became ill with respiratory infections. Tests showed a variety of adenoviruses in the trainees, but at least 106?and probably more?had the mutated form of Ad14, including five who ended up in an intensive care unit
In April, Oregon health officials learned of a cluster of cases at a Portland-area hospital. They ultimately counted 31 cases, including seven who died with severe pneumonia. The next month, Washington state officials reported four hospitalized patients had the same mutated virus. One, who also had AIDS, died.
The Ad14 form of adenovirus was first identified in 1955. In 1969, it was blamed for a rash of illnesses in military recruits stationed in Europe, but it's been detected rarely since then. But it seems to growing more common. The strain accounted for 6 percent of adenovirus samples collected in 22 medical facilities in 2006, while none was seen the previous two years, according to a study published this month in the medical journal Clinical Infectious Diseases.



 38?C (100.5?F)<SUP> </SUP>and a respiratory symptom<SUP> </SUP>(cough or sore throat).<SUP> </SUP>Clinical specimens are obtained<SUP> </SUP>as pharyngeal swabs in<SUP> </SUP>viral transport medium (Remel)<SUP> </SUP>from a subset of<SUP> </SUP>recruits who seek medical<SUP> </SUP>care. These samples are<SUP> </SUP>then sent on dry<SUP> </SUP>ice to the NHRC<SUP> </SUP>to be tested for<SUP> </SUP>respiratory pathogens.<SUP> </SUP>
38?C (100.5?F)<SUP> </SUP>and a respiratory symptom<SUP> </SUP>(cough or sore throat).<SUP> </SUP>Clinical specimens are obtained<SUP> </SUP>as pharyngeal swabs in<SUP> </SUP>viral transport medium (Remel)<SUP> </SUP>from a subset of<SUP> </SUP>recruits who seek medical<SUP> </SUP>care. These samples are<SUP> </SUP>then sent on dry<SUP> </SUP>ice to the NHRC<SUP> </SUP>to be tested for<SUP> </SUP>respiratory pathogens.<SUP> </SUP>

 mol/L<SUP> </SUP>concentration of each primer<SUP> </SUP>instead of 0.2
mol/L<SUP> </SUP>concentration of each primer<SUP> </SUP>instead of 0.2 
 -GTCTTATGTACTATAACAGTACTGG-3
-GTCTTATGTACTATAACAGTACTGG-3

 100 TCID<SUB>50</SUB>) using single<SUP> </SUP>antisera and, as such,<SUP> </SUP>are unlikely to identify<SUP> </SUP>mixtures of multiple serotypes.<SUP> </SUP>
100 TCID<SUB>50</SUB>) using single<SUP> </SUP>antisera and, as such,<SUP> </SUP>are unlikely to identify<SUP> </SUP>mixtures of multiple serotypes.<SUP> </SUP>
Comment